Trajectory SP361
Force field:
martini_v2.2P
Simulation length (ns): 5000
Electric field (kJ mol-1 nm-1 e-1): 0
Temperature (K): 300 (v-rescale)
Pressure (bar): 1 (parrinello-rahman semiisotropic)
Number of particles: 24217
Time step (fs) : 25
Software: GROMACS 2020
Simulation length (ns): 5000
Electric field (kJ mol-1 nm-1 e-1): 0
Temperature (K): 300 (v-rescale)
Pressure (bar): 1 (parrinello-rahman semiisotropic)
Number of particles: 24217
Time step (fs) : 25
Software: GROMACS 2020
Supercomputer:
Finisterrae II CESGA
Peptides: P61 DRAMP01301
Lipids: DOPC, DOPE, DOPS, DPSM
Heteromolecules: CHOL
Ions: NA
Water model: PW
Peptides: P61 DRAMP01301
Lipids: DOPC, DOPE, DOPS, DPSM
Heteromolecules: CHOL
Ions: NA
Water model: PW
Sequence :
FLSLIPHAINAVSAIAKHN
Total charge (e): +1
Number of residues: 19
By amino acid: Basic: 3 Acidic: 0 Hydrophobic: 12 Polar: 4 Electrostatic Dipolar Moment (e nm): 2.19
Longitudinal (e nm): 2.11 Transversal (e nm): 0.59 Hydrophobic Dipolar Moment (nm): 20.7
Longitudinal (nm): 20.2 Transversal (nm): 4.82 Secondary structure: Helix
Activity:
Download Files
ITP file. JSON file. PDB file.
Click on any component to highlight it from the plot.
Upper leaflet
Lower leaflet
Lipids
Membrane model for: DOPC:DOPE:DOPS:DPSM:CHOL (3:3:3:2:4) (Cancer)
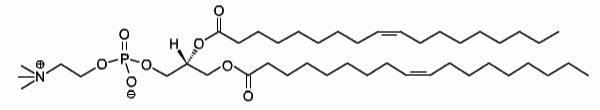
DOPC
1,2-dioleoyl-sn-glycero-3-phosphocholine
Total charge (e): 0
See DOPC lipid
Download ITP File. Download PDB File.
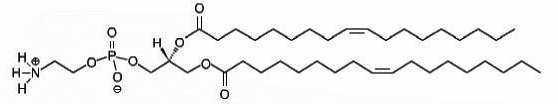
DOPE
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
Total charge (e): 0
See DOPE lipid
Download ITP File. Download PDB File.
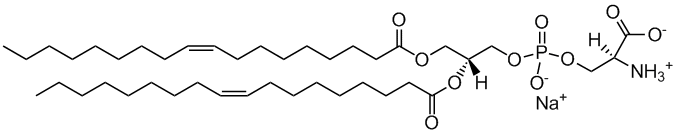
DOPS
1,2-dioleoyl-sn-glycero-3-phospho-L-serine
Total charge (e): -1
See DOPS lipid
Download ITP File. Download PDB File.
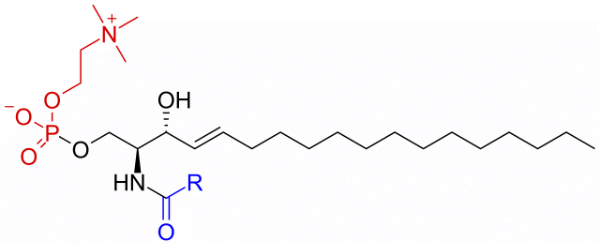
DPSM
Sphingomyelin C(d18:1/18:0) N-stearoyl-D-erythro tails
Total charge (e): 0
See DPSM lipid
Download ITP File. Download PDB File.