Trajectory SP1383
Force field:
martini_v3
Simulation length (ns): 5000
Electric field (kJ mol-1 nm-1 e-1): 0
Temperature (K): 300 (v-rescale)
Pressure (bar): 1 (Parrinello-rahman semiisotropic)
Number of particles: 17401
Time step (fs) : 25
Software: GROMACS 2021.5
Simulation length (ns): 5000
Electric field (kJ mol-1 nm-1 e-1): 0
Temperature (K): 300 (v-rescale)
Pressure (bar): 1 (Parrinello-rahman semiisotropic)
Number of particles: 17401
Time step (fs) : 25
Software: GROMACS 2021.5
Supercomputer:
Finisterrae III CESGA
Peptides: P480 NC00415
Lipids: POPE, POPG
Heteromolecules:
Ions: NA
Water model: W
Peptides: P480 NC00415
Lipids: POPE, POPG
Heteromolecules:
Ions: NA
Water model: W
Sequence :
PGLKGKRGDSGSPATWTTRG
Total charge (e): +3
Number of residues: 20
By amino acid: Basic: 4 Acidic: 1 Hydrophobic: 10 Polar: 5 Electrostatic Dipolar Moment (e nm): 5.37
Longitudinal (e nm): 5.29 Transversal (e nm): 0.9 Hydrophobic Dipolar Moment (nm): 1.28
Longitudinal (nm): 1.26 Transversal (nm): 0.22 Secondary structure: Helix
Activity:
Download Files
ITP file. JSON file. PDB file.
Click on any component to highlight it from the plot.
Upper leaflet
Lower leaflet
Lipids
Membrane model for: POPG:POPE (1:3) (Gram-negative bacteria)
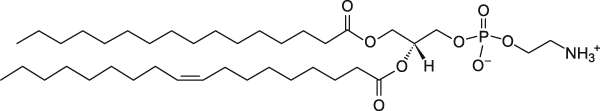
POPE
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
Total charge (e): 0
See POPE lipid
Download ITP File. Download PDB File.
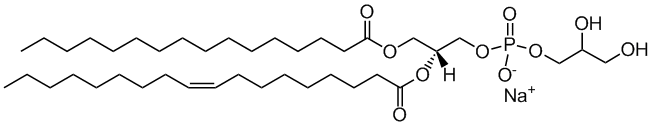
POPG
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol
Total charge (e): -1
See POPG lipid
Download ITP File. Download PDB File.
Last snapshot
Total contacts per residue
Molecular Dynamics based descriptors
Average and standard deviation,
calculated using the autocorrelation function (for time
series)
or the width of the distribution, for the last microsecond
of
the trajectory
Area per lipid
Membrane (nm2): 0.61382800 ± 0.00110239
Upper leaflet (nm2): 0.61382800 ± 0.00110239
Lower leaflet (nm2): 0.61382800 ± 0.00110239
Average Z coordinate
Peptide (nm): 3.641810 ± 0.048308
First Residue (nm): 4.4409700 ± 0.0417992
Last Residue (nm): 2.73720 ± 0.08046
Membrane (nm): 6.4055000 ± 0.0114492
Upper leaflet Head Group (nm): 8.4202700 ± 0.0140128
Lower leaflet Head Group (nm): 4.39018000 ± 0.00921257
Bilayer Thickness (nm): 4.0300900 ± 0.0167699
Peptide insertion (nm): 0.7483690 ± 0.0491786
Contacts
Peptide - Water: 56.850000 ± 0.924071
Peptide - Head groups: 8.342500 ± 0.336635
Peptide - Tail groups: 4.952500 ± 0.206638
Tilt (°): 52.97470 ± 2.22324
Membrane (nm2): 0.61382800 ± 0.00110239
Upper leaflet (nm2): 0.61382800 ± 0.00110239
Lower leaflet (nm2): 0.61382800 ± 0.00110239
Average Z coordinate
Peptide (nm): 3.641810 ± 0.048308
First Residue (nm): 4.4409700 ± 0.0417992
Last Residue (nm): 2.73720 ± 0.08046
Membrane (nm): 6.4055000 ± 0.0114492
Upper leaflet Head Group (nm): 8.4202700 ± 0.0140128
Lower leaflet Head Group (nm): 4.39018000 ± 0.00921257
Bilayer Thickness (nm): 4.0300900 ± 0.0167699
Peptide insertion (nm): 0.7483690 ± 0.0491786
Contacts
Peptide - Water: 56.850000 ± 0.924071
Peptide - Head groups: 8.342500 ± 0.336635
Peptide - Tail groups: 4.952500 ± 0.206638
Tilt (°): 52.97470 ± 2.22324
PepDF:
5(ns): CVS
Displacement (nm): 0.835227 ± 0.035435
Precession(°): 2.29924 ± 4.95623
50(ns) CVS
Displacement (nm): 2.2683100 ± 0.0968232
Precession(°): 25.3791 ± 12.6122
100(ns) CVS
Displacement(nm): 2.81988 ± 0.11968
Precession(°): 45.6961 ± 17.4365
200(ns) CVS
Displacement(nm): 3.361160 ± 0.145684
Precession(°): 103.7200 ± 26.7155
Download JSON File.
5(ns): CVS
Displacement (nm): 0.835227 ± 0.035435
Precession(°): 2.29924 ± 4.95623
50(ns) CVS
Displacement (nm): 2.2683100 ± 0.0968232
Precession(°): 25.3791 ± 12.6122
100(ns) CVS
Displacement(nm): 2.81988 ± 0.11968
Precession(°): 45.6961 ± 17.4365
200(ns) CVS
Displacement(nm): 3.361160 ± 0.145684
Precession(°): 103.7200 ± 26.7155
Download JSON File.
Peptide Analyses
Peptide Displacement Fingerprint
(PepDF)
Lateral displacement vs
Rotational
Displacement along the trajectory, for different time
windows .
Density maps:
2D-density maps of lipids around the
peptide
along XY and YZ axis, calculated for each lipid type along the
last
microsecond.
Lipid-Peptide Analyses:
z-Position
Z-coordinate, averaged for
differetn
parts of the the system: peptide, membrane, first and
last
backbone (BB) residues and upper of lower leaflet
lipids’
headgroups (HGs).
Minimum distance
Minimum distance (nm) between the
peptide backbone and the lipids (headgroups and
tailgroups).
Number of contacts
Number of contacts between the
peptide backbone and the water or the lipids separated
by
lipid headgroups (HG) or lipid tails, using a cut-off of
0.6
nm.
Lateral density
Lateral density for the different
components of the system: headgroups, tail groups,
peptide
and water.